We have now performed experiments in HT29 and HepG2 cells to test mitigation/synthetic lethality between…
Week 3 progress update
Here’s an update on our progress this week:
RNA-seq
- We are actively analyzing and following up on the RNA-seq data we collected last week. We have uploaded the raw data and some processed files run through gene (DESeq2) and pathway (GSEA) analysis tools. While we are actively analyzing these data, we are also already pursuing promising leads.
- Among these leads, the RNA-seq gave clear evidence from all 4 cell lines that HCQ activates cholesterol metabolism genes (see figure below), which is a sign that HCQ is blocking the ability of cells to uptake cholesterol. This phenotype fits with the known role of HCQ in preventing endosome-lysosome trafficking since cholesterol is taken into cells through LDLRs, which are processed through the endosome-lysosome pathway.
- To test whether HCQ does indeed block LDL uptake, we fed fluorescently labeled cholesterol to HepG2 and HT29 cells in the presence or absence of HCQ. In both cell lines, we see robust and significant decrease in LDL uptake (see figure below).
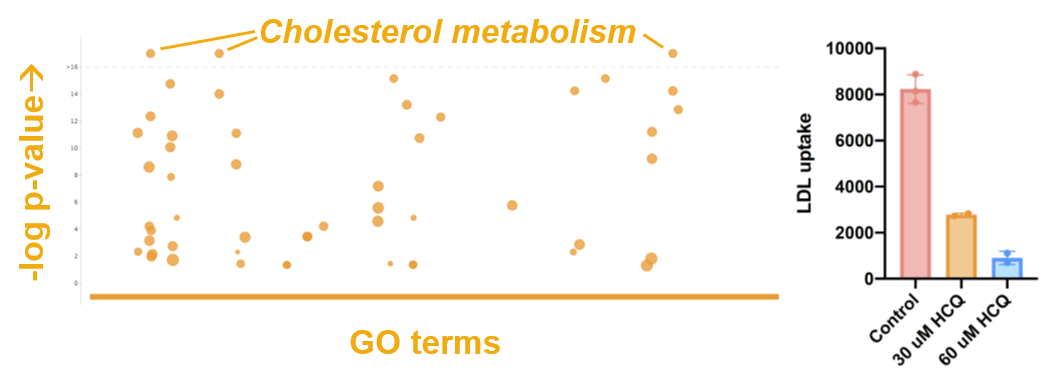
- We are currently asking whether this impairment in cholesterol uptake/processing contributes to HCQ cytotoxicity by asking whether we can exacerbate or mitigate cytotoxicity by further blocking or dosing cells with cholesterol in the presence of HCQ (a synthetic lethality paradigm).
CRISPR screening
- We have completed our drug titration experiments, identifying concentrations of remdesivir and HCQ that yield ~50% survival after 4-6 days of drug treatment. See the table below for specifics.
| Cell line | Concentration of Remdesivir for RNA-seq (<24h), uM | Concentration of HCQ for RNA-seq (<24h), uM | Concentration of Remdesivir for CRISPR screening (4-9 days), uM | Concentration of HCQ for CRISPR screening (4-9 days), uM | Concentration of Puromycin |
| HCT116-Cas9NG-mCherry | 20 | 100 | 2.5 (1:20,000) | 80 (1:625) | 1:20,000 |
| HT29 | 20 | 60 | 1.67 (1:30,000) | 30 (1:1,667) | 1:30,000 |
| HepG2 | 10 | 20 | 1.25 (1:40,000) | 10 (1:5,000) | 1:20,000 |
| PLC/PRF/5 Cas9NG-mCherry | 10 | 80 | 2.5 (1:20,000) | 40 (1:1,250) | 1:20,000 |
- We have now transduced our 4 cell lines with the Brunello genome-wide CRISPR library in lentiCRISPRv2 format in two replicates each. Transduction went smoothly, and now we have added CC50 doses of remdesivir and HCQ (using DMSO as a control) to all cell lines. We plan on harvesting cells by the end of this week for nextgen sequencing.
Establishing a scalable mammalian cell fluorescence-based assay for SARS-CoV2 RdRP activity
- Mia has uploaded a blog post describing the SARS-CoV2 RdRP assay she has designed. We are currently cloning the required components for this assay.
